Introduction
Traditional protein analysis methods are mostly physical and chemical methods. Such as X-ray crystal
diffraction and nuclear magnetic resonance technology. It not only waste a lot of time, but also
consumes a lot of manpower and material resources. Mining the features in the protein sequence
and predicting protein through machine learning can not only greatly improve the prediction efficiency,
but also obtain higher-accuracy results compare with experimental analysis.
The concept of Reduced Amino Acids was proposed in 1960. It has great potential in sequence alignment and structure prediction. In Zuo's article, the PseKRAAC method has been proposed and a web server based on PseKRAAC has been built. In Zheng's web server, 74 types of Reduce Amino Acid Codes and literature sources have been listed in detail.
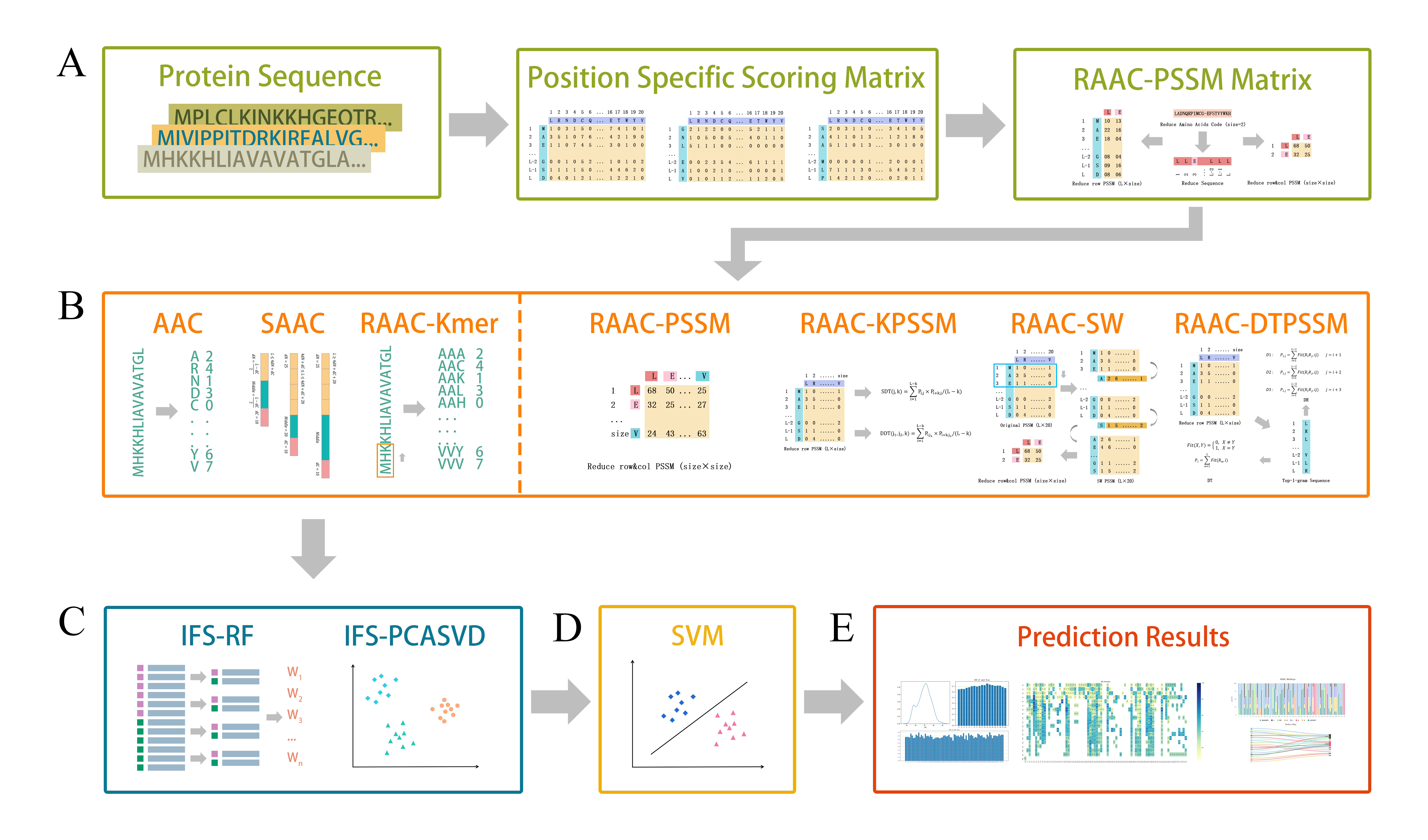
The IRAP toolkit is a dedicated toolkit based on the RAAC-PSSM protein classification prediction method, which developed by Zuo's Lab. It uses 7 feature extraction methods and SVM (Support Vector Machines) for protein classification prediction. You can find almost all common functions which used in protein classification in the IRAP toolkit, and get better classification models and more diverse results analysis.
The concept of Reduced Amino Acids was proposed in 1960. It has great potential in sequence alignment and structure prediction. In Zuo's article, the PseKRAAC method has been proposed and a web server based on PseKRAAC has been built. In Zheng's web server, 74 types of Reduce Amino Acid Codes and literature sources have been listed in detail.
The IRAP toolkit is a dedicated toolkit based on the RAAC-PSSM protein classification prediction method, which developed by Zuo's Lab. It uses 7 feature extraction methods and SVM (Support Vector Machines) for protein classification prediction. You can find almost all common functions which used in protein classification in the IRAP toolkit, and get better classification models and more diverse results analysis.