Help
OVERVIEW
Introduction
It is a new sequence logo generator by using reduced amino acid alphabets. Users can easily generate many differ-ent simplified amino acid weblogos tailored to their needs by selecting various reduced amino acids alphabets within underlying biology knowledges. This current webserver provide 74 types of reduced amino acid alphabet, which were manually extracted to generate 673 reduced amino acid clusters (RAACs) for dealing with protein alignment. A two-dimensional box was proposed for easily selecting desired RAACs with underlying biology knowledges. It is antici-pated that the RaacLogo web server will play more high-potential roles for protein sequence alignment, topological estimation and protein design experiments.Feedback
We welcome any feedback. If you find errors, omissions, or if you want to suggest new RAACs or applications being added to RaacLogo, please let us know. You can contact us by Email (yczuo@imu.edu.cn, baimoc@163.com).ANALYSIS
Step1. Enter Query Sequence
User firstly go to RaacLogo homepage (http://bioinfor.imu.edu.cn/RaacLogo), and paste the query protein sequence with equal length FASTA format. There are two ways supported, the one is to upload file with amino acid sequences, the other is to paste sequences into the input box directly.Step2. RAAC Type Selection
Users can select the desired RAAC type in a two-dimensional box, which displays the different alphabet types and cluster sizes. If users want to know more about RAAC type, they can click on the type id to enter the information page on the box left. This will show the type name, description, clustering details and reference. The user also can conveniently select all the clusters of the single type, and click the analysis button to generate all RaacLogo about this type.
If interested reduced cluster is not found in the presented box option, we also supply the user-defined choice. Just needing to enter reduced alphabet derived from user own definition in the input box named "Custom RAAC", the user-defined RaacLogo will be generated. It is worth noting that we need the 20 amino acids in the single letter, and use "," as the separator to distinguish each cluster.
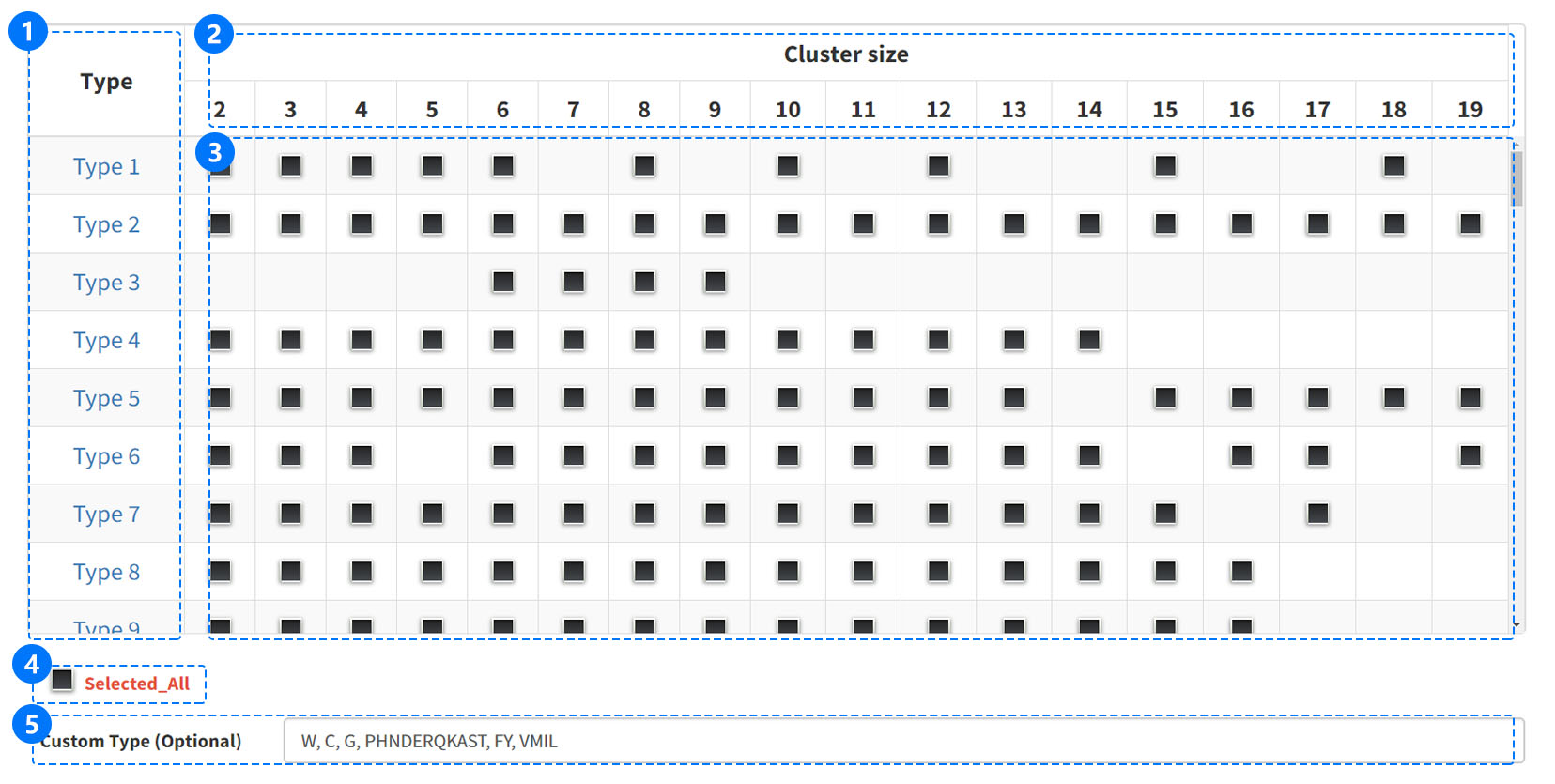
(1) In the "Type" column, each number represents an amino acid alphabet reduction type. If you hover mouse over the type, will get a brief description of type.However, if you click on the type, you will be taken to the information page.
(2) In the "Cluster size" column, the row shows all clustering information, and the number indicates cluster size.
(3) Click the gray button to select the cluster and the button will change to blue.
(4)
Click the "Selected All" button, at the bottom of the box two-dimensional selection box, to select all the RAAC types.
(5)
The custom type can to group the 20 amino acids into any number of clusters arbitrarily as new reduction scheme.
3. Result Output
The websever can generates up to more than 670 RaacLogos based on discrete reduction rules in one analysis job. Each line repre-sents a logo report generated by RAAC, which contains job id, the details of the RAAC, the natural logo and RaacLogo.Reduced logo report displays three results of sequence logo, The first line gives the regular logo from 20 natural amino acids. The second line is the RaacLogo replaced by using the first letter of each reduced subcluster. For example, if the 20 natural amino acids are clustered based on hydrophobicity classification (Type 54), there will be three reduced subclusters obtained. They are Polar (RKEDQN), Neutral (GASTPHY), Hydrophobicity (CLVIMFW), respectively. And the second line gives RaacLogo containing three represented letters of reduced amino acids by using R, G, and C. The third line gives the logo by using only color representation, one color shows these amino acids are the same subcluster. All RaacLogo provides png, svg, pdf formats for downloading.
i. Download List
Each RAAC types provide corresponding fasta, csv and libsvm vector files for download.All these files are compressed into a zip file.
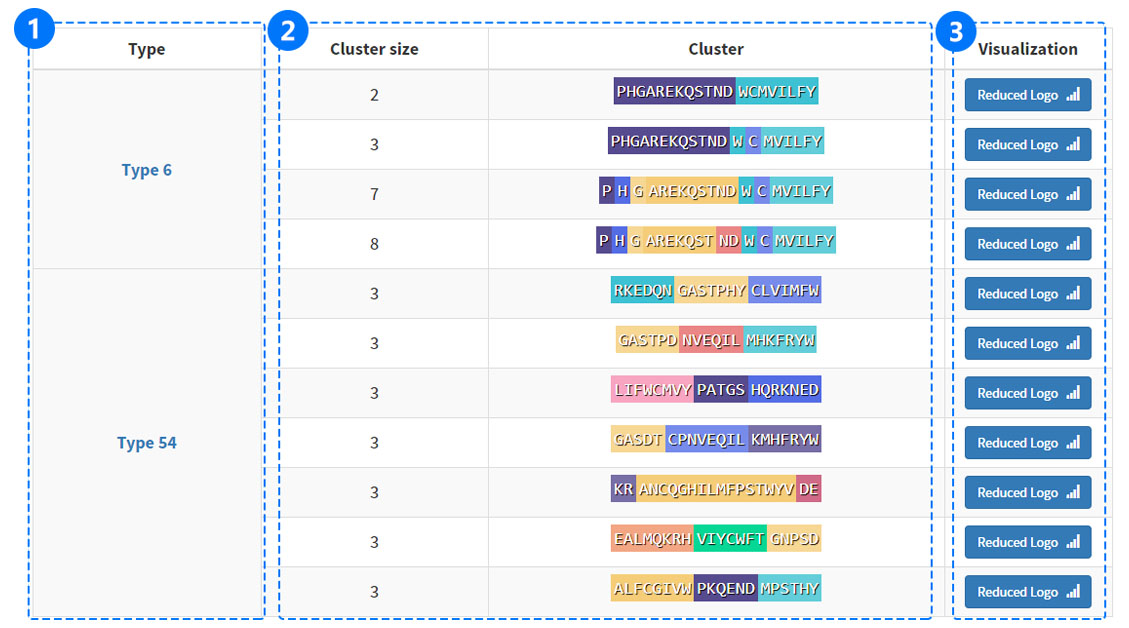 (1) Click the types to visit INFORMATION.
(1) Click the types to visit INFORMATION.
(2) The reduced amino acids cluster (RAAC) are shown as a bar with amino acid residues of different colors.
(3) Enter page of RaacLogo.
iv. RaacLogo
 (1) Amino acid distribution of each site of the natural sequence, based on the alphabet with 20 amino acids;
Amino acid distribution of each site of the reduced sequence, based on the reduced amino acid clusters.
(1) Amino acid distribution of each site of the natural sequence, based on the alphabet with 20 amino acids;
Amino acid distribution of each site of the reduced sequence, based on the reduced amino acid clusters. (2) Download three files (svg, pdf, png) from grey button for future research.
INFORMATION
 The type id, name, description and other are showed in the information page. It is important that the amino acid reduction clusters for each method are visualized by clustering with different colors. Users can easily analyze the selected RAAC by clicking "Analysis" button to enter the ANALYSIS.
The type id, name, description and other are showed in the information page. It is important that the amino acid reduction clusters for each method are visualized by clustering with different colors. Users can easily analyze the selected RAAC by clicking "Analysis" button to enter the ANALYSIS.
(1) The information includes selected type, name, description and reduced amino acids cluster.
(2) The reduced amino acids cluster (RAAC) are shown as a bar with amino acid residues of different colors.
(3) Click the button to visit the analysis interface with selected RAAC types.